
Received: NovemAccepted: FebruPublished: March 31, 2011Ĭopyright: © 2011 Thompson et al. Craig Venter Institute, United States of America PLoS ONE 6(3):Įditor: Jonathan Badger, J. We then propose knowledge-enabled, dynamic solutions that will hopefully pave the way to enhanced alignment construction and exploitation in future evolutionary systems biology studies.Ĭitation: Thompson JD, Linard B, Lecompte O, Poch O (2011) A Comprehensive Benchmark Study of Multiple Sequence Alignment Methods: Current Challenges and Future Perspectives. Based on this study, we demonstrate that the existing MSA methods can be exploited in combination to improve alignment accuracy, although novel approaches will still be needed to fully explore the most difficult regions. Third, the badly predicted or fragmentary protein sequences, which make up a large proportion of today's databases, lead to a significant number of alignment errors. Second, motifs in natively disordered regions are often misaligned.
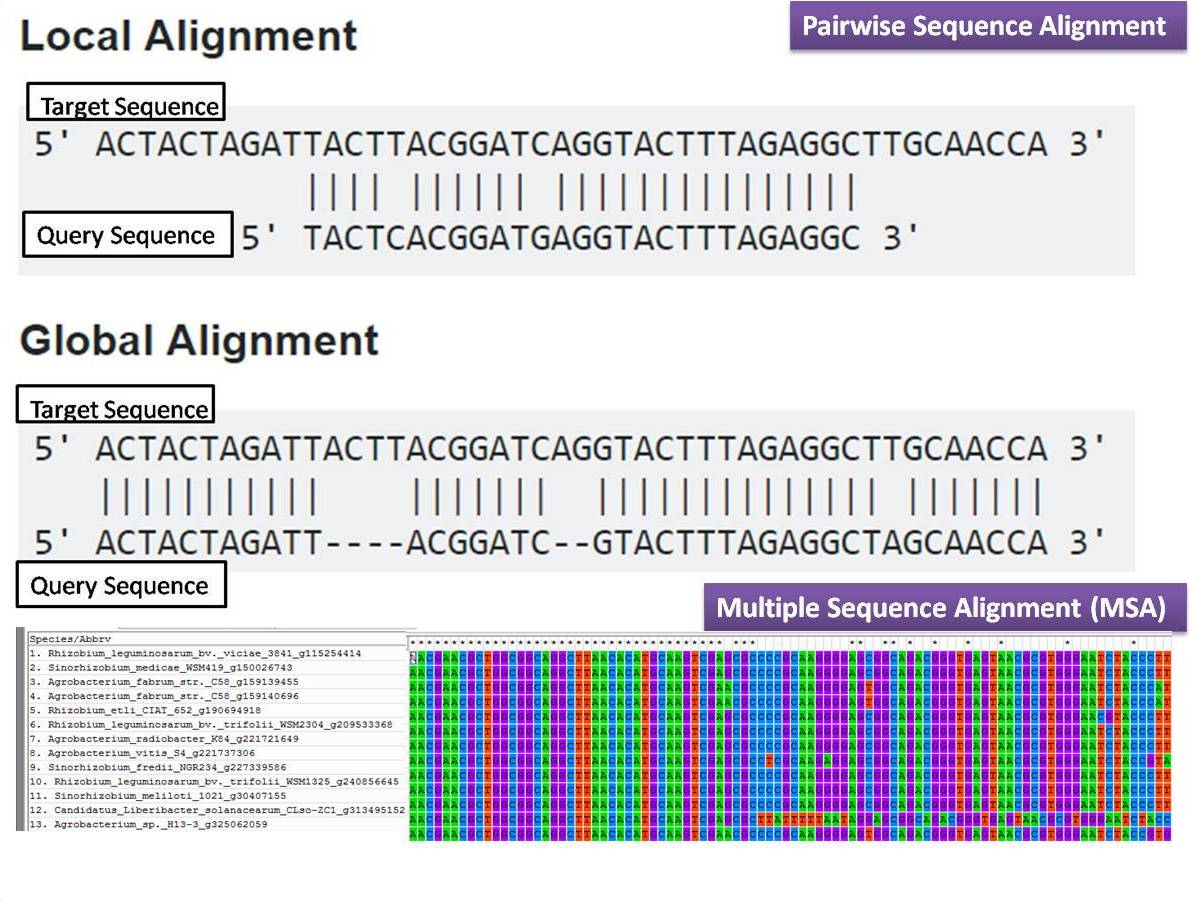
First, the locally conserved regions, that reflect functional specificities or that modulate a protein's function in a given cellular context,are less well aligned. However,we have identified a number of important challenges. We show that alignmentmethods have significantly progressed and can now identify most of the shared sequence features that determine the broad molecular function(s) of a protein family, even for divergent sequences. The benchmark is designed to represent typical problems encountered when aligning the large protein sequence sets that result from today's high throughput biotechnologies. In this paper, we describe a comprehensive evaluation of many of the most popular methods for multiple sequence alignment (MSA), based on a new benchmark test set.
By placing the sequence in the framework of the overall family, multiple alignments can be used to identify conserved features and to highlight differences or specificities. The base editing system, capable of achieving the precise editing of DNA or RNA, is considered a transformative technology for gene function research, disease treatment, and biological breeding.Multiple comparison or alignmentof protein sequences has become a fundamental tool in many different domains in modern molecular biology, from evolutionary studies to prediction of 2D/3D structure, molecular function and inter-molecular interactions etc. The development of a high-throughput protein clustering method based on a three-dimensional structure will give protein function research support, and promote the function mining of unknown proteins, since the protein function is determined by its three-dimensional spatial structure. The results showed that the discovered deaminases, based on AI-assisted structural predictions, greatly expands the utility of base editors for therapeutic and agricultural applications. They developed the novel base editing system and tested it in animal and plant cells.

The researchers from the Chinese Academy of Sciences predicted the three-dimensional structure of proteins with deamination functional sequences and carried out multiple alignment and clustering of proteins. BEIJING, July 2 (Xinhua) - Chinese researchers have built a novel protein clustering method and developed a new base editing system, supporting protein function analysis and new functional element exploration, according to a recent research article published in the journal Cell.įunctional clustering of proteins is an important method to understand their participation in physiological processes and to design new proteins.


 0 kommentar(er)
0 kommentar(er)
